Publications
2026
Impact of Lipid Composition on Membrane Partitioning and Permeability of Gas Molecules.
Membranes, 16: 33 (cover page).
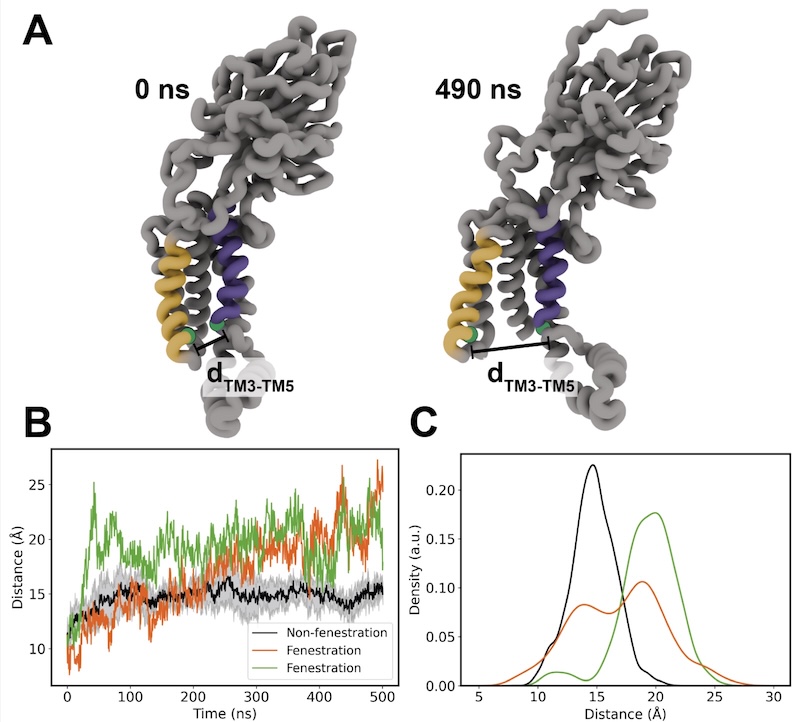
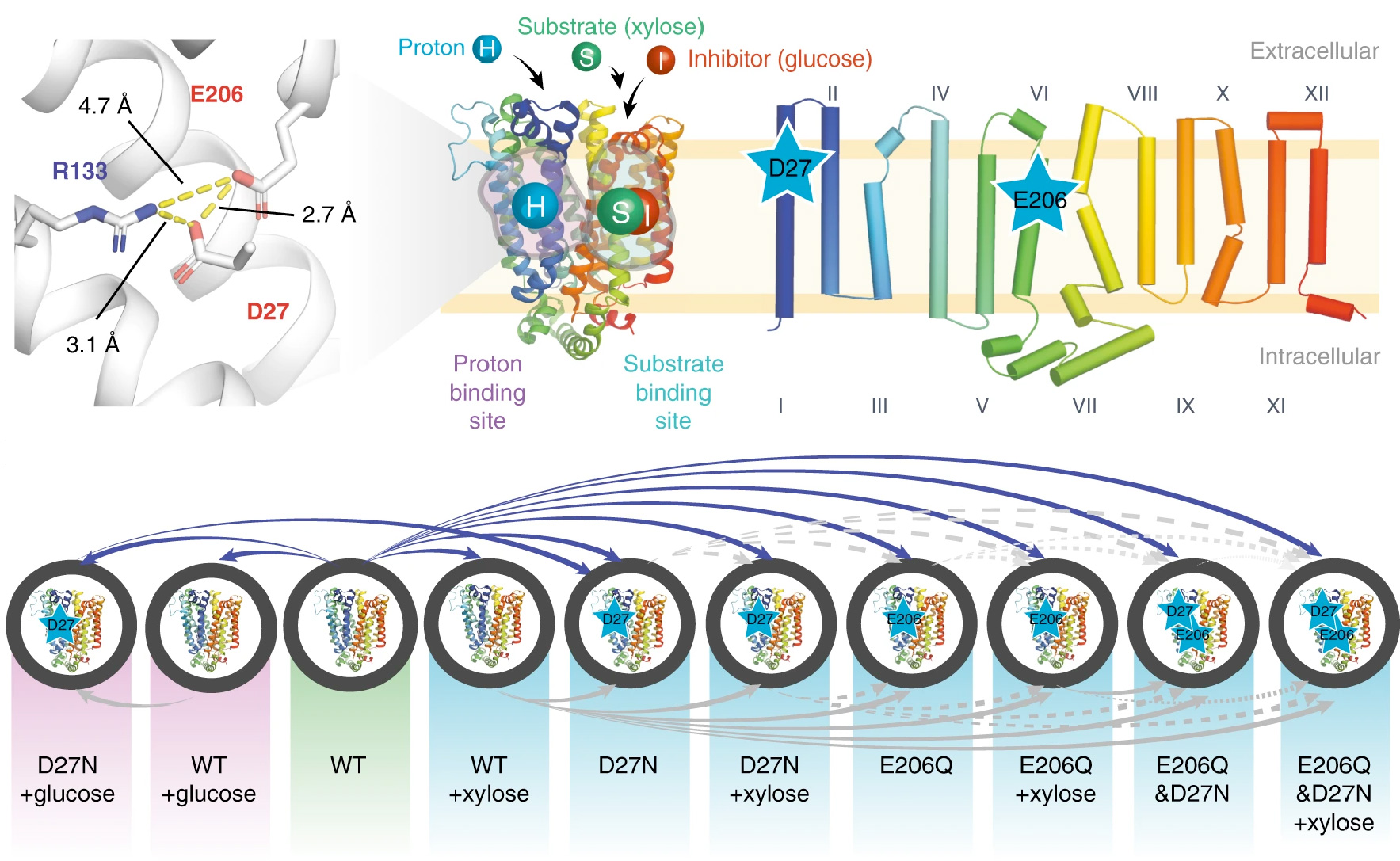
Elevator mechanism dynamics in a sodium-coupled dicarboxylate transporter.
PNAS, 123: e2500723123.
Thermodynamic and Kinetic Analysis of Molecular Conformational Dynamics in a Riemannian Framework.
JPCA, in press.
ABCA1 activity in myeloid immune cells results in an anti-cancer phenotype.
Science Advances, in press.
BioRXiv: https://www.biorxiv.org/content/10.1101/2025.02.19.638515v1
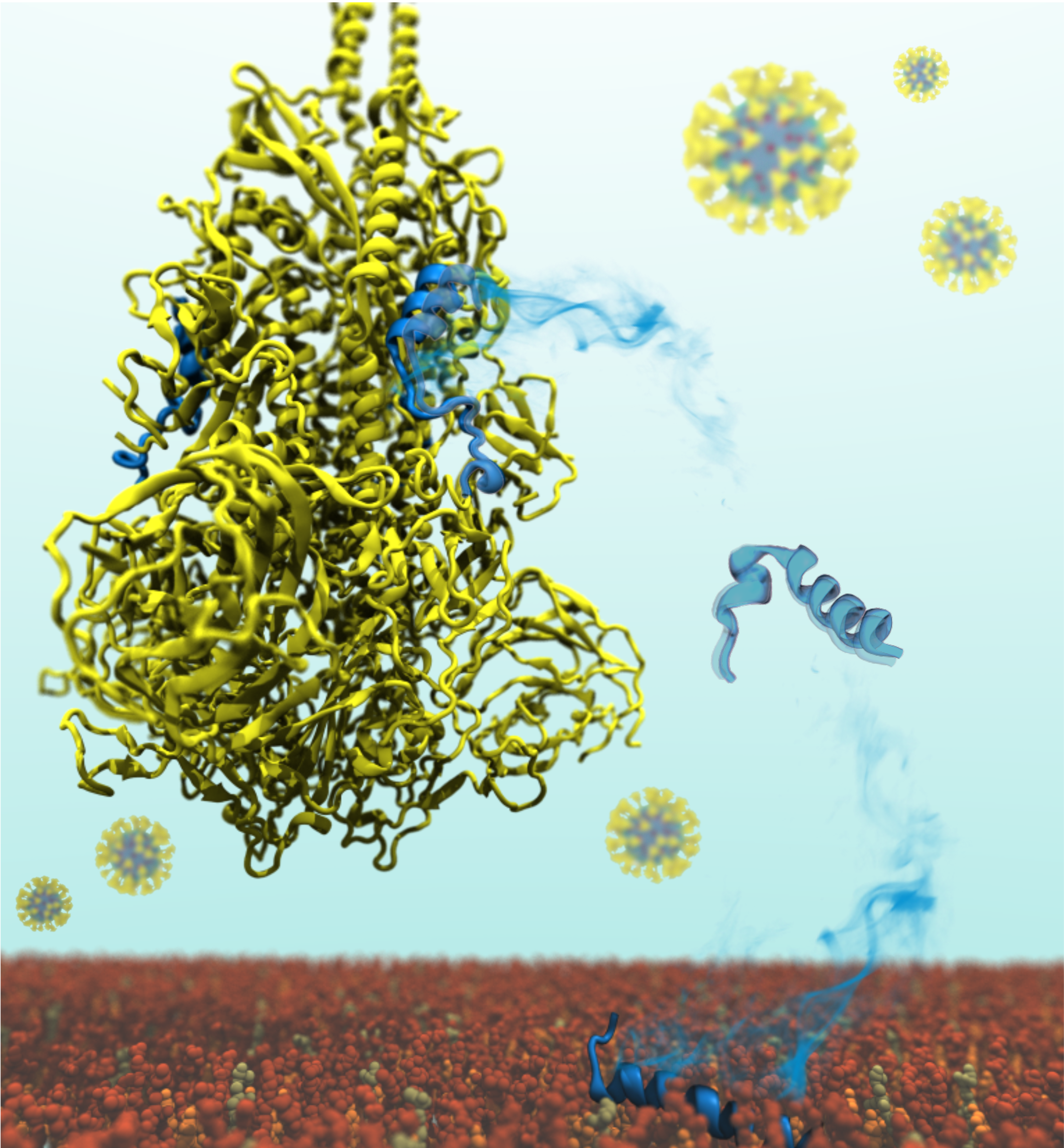
Probing the Role of Membrane in Neutralizing Activity of Antibodies Against Influenza Virus.
Structure, in press.
( Free Download Till 07 Feb 2026)
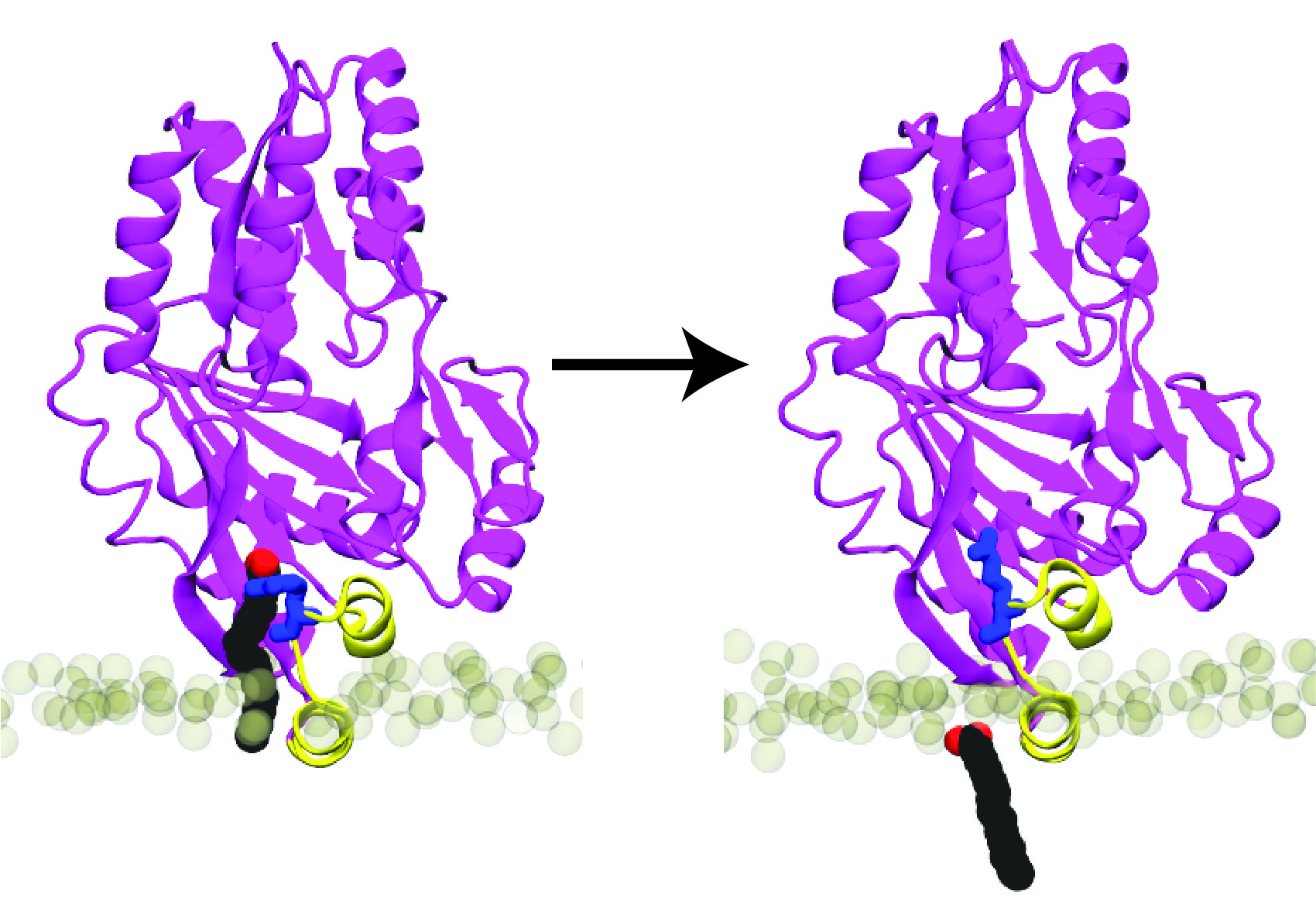
E. coli LetA defines a structurally distinct transporter family involved in lipid trafficking.
Nature, in press.
Structure of nanobody-inhibited 1 state of human bile salt transporter NTCP.
Structure, in press.
Simulating gas permeation through the central pore of AQP5.
Advances in Experimental Medicine and Biology, in press.
Characterization of membrane binding and protein-lipid interactions at atomic level with accelerated HMMM model.
Methods in Molecular Biology, in press.
2025
Conformational free energy landscape of a glutamate transporter and microscopic details of its transport mechanism.
PNAS, 122, e2416381122.
Impact of anionic lipids on the energy landscape of conformational transition in anion exchanger 1 (AE1).
Nature Communications, 16: 11664.
Ca2+ stoichiometry controls the binding mode of PKCα C2 domain to anionic membranes.
J. Phys. Chem. B, 129: 9893-9903.
Dissecting Large-Scale Structural Transitions in Membrane Transporters using Advanced Simulation Technologies.
Perspective Article, J. Phys. Chem. B, 129: 3703–3719 (cover article).
Membrane-bound model of the ternary complex between factor VIIa/tissue factor and factor X.
Blood Advances, 9: 729-740. (cover article).
An orchestrated interaction network at the binding site of human SERT enables the serotonin occlusion and import.
Biochemistry, 64: 3652−3662.
Molecular dynamics simulations of biological membrane systems across scales.
Current Opinion in Structural Biology, 93: 103071.
( Free Download Till 11 Feb 2026)
Voltage Sensor Conformations Induced by LQTS-associated Mutations in hERG Potassium Channels.
Nature Communications, 16, 7126.
Drug-bound Outward-facing Conformation of a Heterodimeric ABC Exporter Suggests a Putative Mechanism of Drug Translocation.
Nature Communications, 16, 10403..
Ca2+ Single-molecule imaging reveals the role of membrane-binding motif and C-terminal domain of RNase E in its localization and diffusion in Escherichia coli.
eLife, 14: RP105062.
Restricted surface diffusion of cytochromes on bioenergetic membranes with anionic lipids.
Membranes, 15: 124. (cover article)
Regulation of Rho guanine nucleotide exchange factor 3 (ARHGEF3) by phosphorylation in the PH domain.
iScience, 28: 112753.
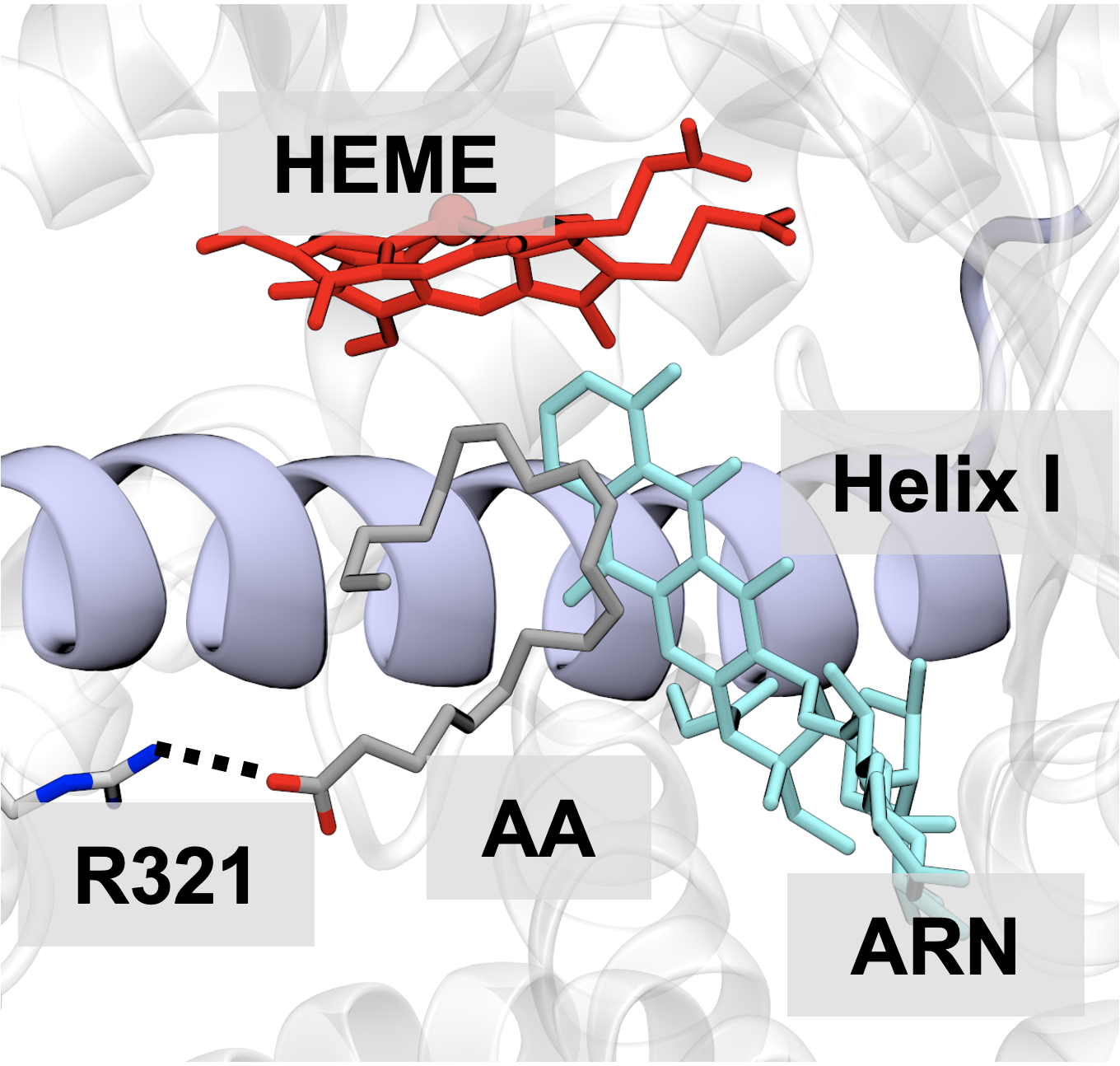
Amino Acid Sequence Controls Enhanced Electron Transport in Heme-Binding Peptide Monolayers.
ACS Central Science, 11: 512-621.
Modeling diffusive motion of ferredoxin and plastocyanin on the PSI domain of Procholorococcus marinus MIT9313.
J. Phys. Chem. B, 129: 52-70.
The String Method with Swarms of Trajectories: A Tutorial for Free-Energy Calculations Along a Zero-Drift Pathway.
J. Phys. Chem. B, 129: 6837-6846..
Cryo-EM structure of the tissue factor/factor VIIa complex with a factor X mimetic reveals a novel allosteric mechanism.
Blood, 146: 2833-2842. (cover article).
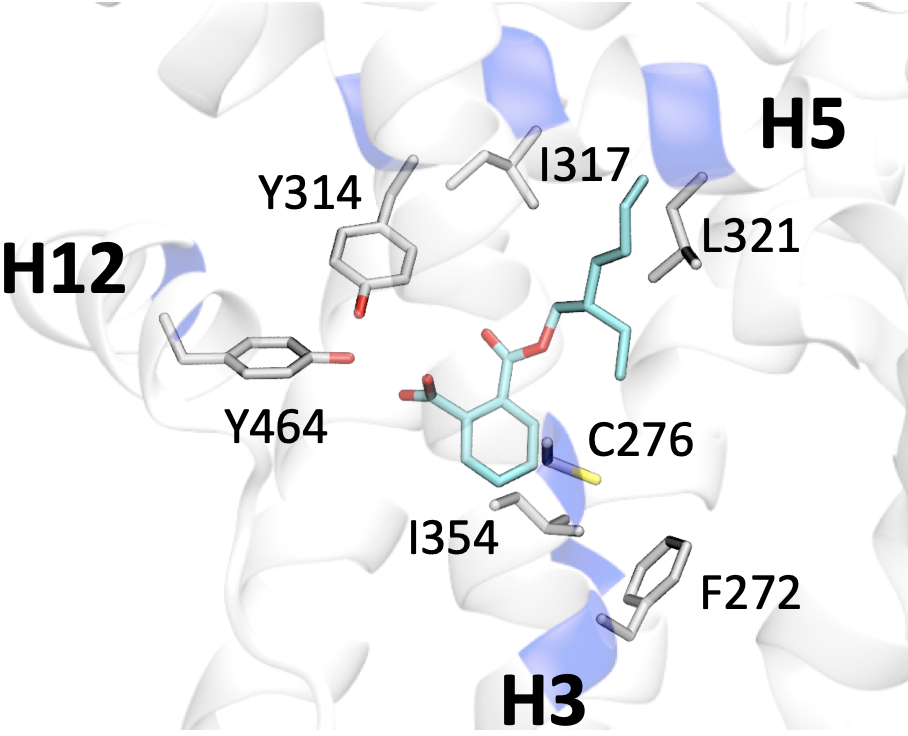
Cholesterol-targeting Wnt/β-catenin signaling inhibitors for colorectal cancer.
Nature Chem. Biol., 21: 1376-86.
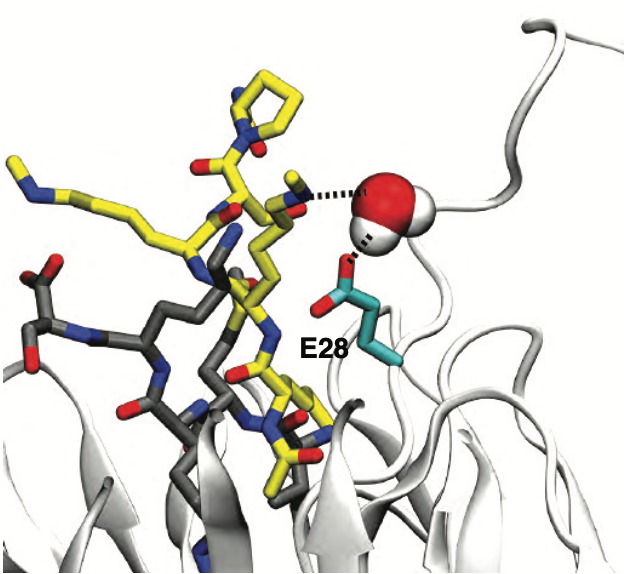
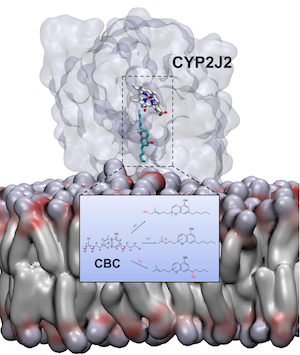
Distinct Interactions of Cannabinol and Its Cytochrome P450-Generated Metabolites with Receptors and Sensory Neurons.
J. Med. Chem., 68: 13935-13953.
2024
GOLEM: Automated and Robust Cryo-EM-guided Ligand Docking with Explicit Water Molecules.
JCIM, 64: 5680–5690.
Generating Concentration Gradients across Membranes for Molecular Dynamics Simulations of Periodic Systems.
Int. J. Mol. Sci., 25: 3616.
Improved Highly Mobile Membrane Mimetic Model for Investigating Protein-Cholesterol Interactions.
JCIM, 64: 4822–4834.
A generative artificial intelligence framework based on a molecular diffusion model for the design of metal–organic frameworks for carbon capture.
Communications Chemistry, 7:21. (cover page)
Topological Learning Approach to Characterizing Biological Membranes.
JCIM, 64: 5242–5252.
Atomistic Characterization of Beta-2-Glycoprotein I Domain V Interaction with Anionic Membranes.
J. Thromb. Hemost., 22: 3277-3289.
The Role of Protein-Lipid Interactions in Priming the Bacterial Translocon.
Membranes, 14: 249.
Secondary structure determines electron transport in peptides.
PNAS, 121: e2403324121.
A Gram-negative-selective antibiotic spares the gut microbiome and prevents Clostridioides difficile infection.
Nature, 630: 429–436.
Tumor-acquired somatic mutation blocks ABCG2 conformational switch required for transport.
Drug Resistance Updates, 73: 101066.
Structure of the human dopamine transporter and mechanisms of allosteric inhibition.
Nature, 362: 672-677.
The carboxy terminus causes interfacial assembly of oleate hydratase on a membrane bilayer.
J. Biol. Chem., 300: 105627.
Molecular mechanisms of functional impairment for active site mutations in glucose-6-phosphatase catalytic subunit 1 linked to glycogen storage disease type 1a.
PNAS Nexus, 3: 306.
ACS Chem. Biol., 19: 2304-2313.
WSB1/2 target chromatin-bound lysine-methylated RelA for proteasomal degradation and NF-κB termination.
Nucleic Acid Research, 52: 4969-4984.
Structure of alpha-synuclein fibrils derived from human Lewy body dementia tissue.
Nature Communications, 15: 2750.
Elucidating the Mechanism of Metabolism of Cannabichromene (CBC) by Human Cytochrome P450s.
J. Nat. Products, 87: 639–651.
Beyond Nothingness: Formation and Functional Relevance of Voids in Polymer Films.
Nature Communications, 15:2852.
Broadening access to small-molecule parameterization with the Force Field Toolkit.
J. Chem. Phys., 160, 242501.
IHMCIF: An extension of PDBx/mmCIF data standard for integrative structure determination methods.
J. Mol. Biol., 436: 168546.
Open Access
2023
VMD as a Platform for Interactive Small Molecule Preparation and Visualization in Quantum and Classical Simulations.
J. Chem. Info. Modeling, 63: 4664–4678.
Targeting lipid-protein interaction for drug development: Development of a novel resistance-proof Syk inhibitor for acute myeloid leukemia.
Nat. Chem. Biol., 19: 239–250.
Tuning aromatic contributions by site-specific encoding of fluorinated phenylalanine residues in bacterial and mammalian cells.
Nature Communications, 14: 59.
Capturing the interplay of membrane lipids and structural transitions in human ABCA7.
EMBO J., 42, e111065.
Structures and membrane interactions of native serotonin transporter in complexes with psychostimulants.
PNAS, 120 (29), e2304602120.
Insights into substrate transport and water permeation in the mycobacterial transporter MmpL3.
Biophys. J., 122: 2342–2352.
300. H. Park, R. Zhu, E. A. Huerta, S. Chaudhuri, E. Tajkhorshid, and D. Cooper (2023)
End-to-end AI framework for interpretable prediction of molecular and crystal properties.
Machine Learning: Science and Technology, 4: 025036.
Quinoline-based Zinc Ionophores with Antimicrobial Activity.
J. Med. Chem., 66: 11078−11093 (cover article)
Asymmetric conformations and lipid interactions shape the ATP-coupled cycle of a heterodimeric ABC transporter.
Nature Communications , 14: 7184
BioRXiv
A computational spatial whole-cell model for hepatitis B viral infection and drug interactions.
Scientific Reports, 13: 21392
296. Y. Zhang, O. Soubias, S. Pant, F. Heinrich, A. Vogel,
J. Li, Y. Li, L. A. Clifton, S. Daum, K. Bacia,
D. Huster, P. A. Randazzo, M. Lösche, E. Tajkhorshid,
and R. A. Byrd (2023)
Myr-Arf1 conformational flexibility at the membrane surface: insights for ArfGAP ASAP1 interactions.
Nature Communications , 14:7570.
A rigorous framework for calculating protein-protein binding affinities in membranes.
JCTC, 19: 9077–9092
294. S. Marru, M. Pierce, B. Plale, S. Pamidighantam, D. Wannipurage, M. Christie,
I. Ranawaka, E. Abeysinghe, R. Quick, E. Tajkhorshid, S. Koric, J. Basney,
M. Spivak, B. Isralewitz, R. Bernardi, D. Gomes, G. Krishnan, M. Bazhenov,
S. Smallen, A. Majumdar, A. Arkhipov, K. Dai and X.-P. Liu (2024)
Cybershuttle: An End-to-end Cyberinfrastructure Continuum to Accelerate Discovery in Science and Engineering.
PEARC’23, July 2023, Pages 26–34.
2022
Structure of C. elegans TMC-1 complex illuminates auditory mechanosensory transduction.
Nature, 610: 796–803.
Lipid-mediated organization of prestin in the outer hair cell membrane and its implications in sound amplification.
Nature Communications, 13:6877.
Post-Translational Modifications Optimize the Ability of SARS-CoV-2 Spike for Effective Interaction with Host Cell Receptors.
PNAS, 119: e2119761119.
Proton-driven alternating access in a spinster transporter, an emerging family of broad-specificity efflux pumps.
Nature Communications, 13: 5161.
Role of internal loop dynamics in antibiotic permeability of outer membrane porins.
PNAS, 119 (8), e2117009119.
Differential dynamics and direct interaction of bound ligands with lipids in multidrug transporter ABCG2.
PNAS, 120: e2213437120.
Assembly and Analysis of Cell-Scale Membrane Envelopes.
J. Chem. Inf. Modeling, 62, 602–617. (cover article)
Microscopic Characterization of the Chloride Permeation Pathway in the Human Excitatory Amino Acid Transporter 1 (EAAT1).
ACS Chem. Neuroscience, 13: 776–785 (cover article).
Membrane Mixer: a toolkit for efficient shuffling of lipids in heterogeneous biological membranes.
J. Chem. Inf. Modeling, 62: 986-996. (cover article)
Anionic lipids confine cyt. c2 to the surface of bioenergetic membranes without compromising its interaction with redox partners.
Biochemistry, 61: 385-397. (cover article)
Extended-Ensemble Docking to Probe Dynamic Variation of Ligand Binding Sites During Large-Scale Structural Changes of Proteins.
Chem. Sci., 13: 4150-4169.
py-MCMD: Python Software for Performing Hybrid Monte Carlo/Molecular Dynamics Simulations with GOMC and NAMD.
J. Chem. Theo. Comp., 18: 4983-94. (cover article)
Molecular View into Preferential Binding of the Factor VII Gla Domain to Phosphatidic Acid.
Biochemistry, 16:1694-1703.
280. L. Leisle, K. Lam, S. Dehghani-Ghahnaviyeh, E. Fortea, J. Galpin, C.
A. Ahern, E. Tajkhorshid, and A. Accardi (2022)
Backbone amides are key determinants of Cl− selectivity in CLC ion channels.
Nature Communications, 14: 59.
279. D. J. Hardy, J.Choi, W. Jiang, and E. Tajkhorshid (2022)
Experiences porting NAMD to the Data Parallel C++ programming model.
International Workshop on OpenCL (IWOCL’22), Association for Computing Machinery, New York, USA. May 2022, 15: 1-5.
Ataxia-linked SLC1A3 mutations alter EAAT1 chloride channel activity and glial regulation of CNS function.
J. Clin. Inv., 132(7): e154891.
Companion Guide to the String Method with Swarms of Trajectories: Characterization, Performance, and Pitfalls.
J. Chem. Theo. Comp., 18: 1406-22.
Anthracycline Derivatives Inhibit Cardiac CYP2J2.
J. Inorg. Biochem., 229, 111722.
Identification of Structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J. Biol. Chem., 298: 101676.
Phthalate Metabolites Act Through Peroxisome Proliferator-Activated Receptors (PPAR) in the Mouse Ovary.
Reproductive Toxicology, 110:113-123.
273. A. Trifan*, D. Gorgun*, M Salim, Z. Li, A. Brace, M. Zvyagin, H. Ma, A. Clyde,
D. Clark, D. J. Hardy, T. Burnley, L. Huang, J. McCalpin, M. Emani, H. Yoo, J. Yin,
A. Tsaris, V. Subbiah, T. Raza, J. Liu, N. Trebesch, G. Wells, V. Mysore, T. Gibbs,
J. C. Phillips, S. C. Chennubhotla, I. Foster, R. Stevens, A. Anandkumar, V. Vishwanath,
J. E. Stone, E. Tajkhorshid, S. A. Harris, and A. Ramanathan (2022)
Intelligent Resolution: Integrating cryo-EM with AI-driven multi-resolution
simulations to observe the SARS-CoV-2 replication-transcription machinery in action.
Int. J. High-Performance Computing
Applications, 36 (5-6): 603-623. .
272. R. P. Sparks*, A. Becker*, A. S. Arango*, E. Tajkhorshid, R. A. Fratti (2022)
Use of Microscale Thermophoresis to Measure Protein-Lipid Interactions.
J. Vis. Exp., 180: e60607.
2021
Molecular Mechanism of Capsid Disassembly in Hepatitis B Virus. PNAS, 118, e2102530118.
Glutamate transporters have a chloride channel with two hydrophobic gates.
Nature, 591, 327-331.
Rationalizing generation of broad spectrum antibiotics with the addition of a positive charge.
Chemical Science, 12, 15028-15044. (Cover Article)
Cell, 184: 4669-4679.e13.
PIP2-dependent coupling of voltage sensor and pore domains in Kv7.2 channel.
Communications Biology, 4, 1189.
Binding Mode of SARS-CoV2 Fusion Peptide to Human Cellular Membrane.
Biophys. J., 120, 2914–2926.
BioRxiv preprint
Defining the Energetic Basis for a Conformational Switch Mediating Ligand-Independent Activation of Mutant Estrogen Receptors in Breast Cancer.
Molecular Cancer Research, 19: 1559-1570 (cover article).
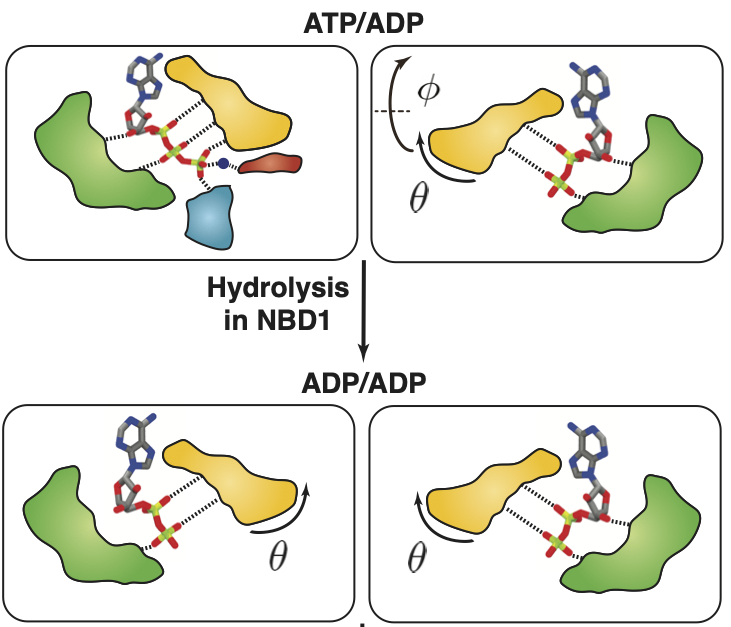
Conformational changes of the nucleotide binding domains of P-glycoprotein induced by ATP hydrolysis.
FEBS Letters, 595: 735-749.
Structural Basis of Complex Formation Between Mitochondrial Anion Channel VDAC1 and Hexokinase-II.
Communications Biology, 4:667.
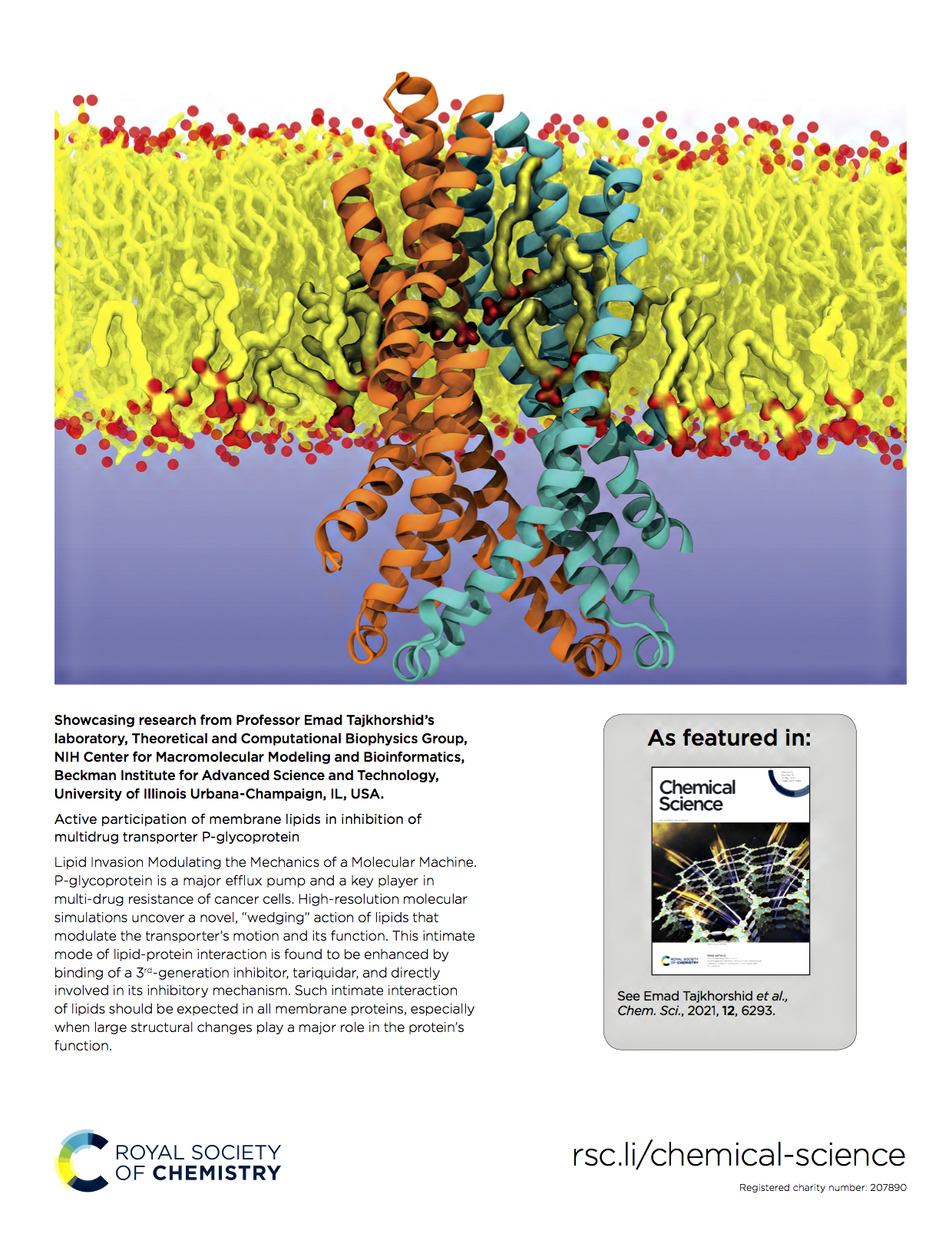
Active Participation of Membrane Lipids in Inhibition of Multidrug Transporter P-Glycoprotein.
Chemical Science, 12: 6293-6306. Featured Article.
261. C. L. Brooks III, D. A. Case, S. Plimpton, B. Roux, D. van der Spoel,
and E. Tajkhorshid (2021)
Classical molecular dynamics.
Editorial Introduction to the JCP Special Topic on Classical Molecular Dynamics (MD) Simulations:
Codes, Algorithms, Force Fields, and Applications.
J. Chem. Phys., 154, 10041.
260. D. J. Hardy, J. E. Stone, B. Isralewitz, and E. Tajkhorshid (2021)
Lessons Learned from Responsive Molecular Dynamics Studies of the COVID-19 Virus.
IEEE/ACM HPC for Urgent Decision Making (UrgentHPC) 1-10.
CO2 Transport across Membranes.
Interface Focus, 11: 20200090.
Amphiphilic Distyrylbenzene Derivatives as Potential Therapeutic and Imaging Agents for Soluble and Insoluble Amyloid β Aggregates in Alzheimer’s Disease.
JACS, 143: 10462–10476.
257. W. R. Arnold, L. N. Carnevale, Z. Xie, J. L. Baylon, E. Tajkhorshid, H. Hu, and A. Das (2021)
Anti-inflammatory dopamine- and serotonin-based endocannabinoid epoxides reciprocally regulate cannabinoid receptors and the TRPV1 channel.
Nature Communications, 12, 926.
Cryo-EM structures of E. coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site. PNAS, 118, e2106750118.
255. J. Zabret, S. Bohn, S. K. Schuller, O. Arnolds, M. Möller, J. Meier-Credo,
P. Liauw, A. Chan, E. Tajkhorshid, J. D. Langer, R. Stoll, A. Krieger-Liszkay,
B. D. Engel, T. Rudack, J. M. Schuller, and M. M. Nowaczyk (2021)
How to build a water-splitting machine: structural insights into photosystem II assembly.
Nature Plants, 7: 524–538.
CryoFold: determining protein structures and data-guided ensembles from cryo-EM density maps.
Matter, 4: 3195-3216.
Cation-π interactions and their functional roles in membrane proteins.
Journal of Molecular Biology, 433: 167035.
252. W.-M. Kwok, E. Tajkhorshid, and A. K. S. Camara (2021)
Editorial: Mitochondrial Exchangers and Transporters in Cell Survival and Death.
Frontiers in Physiology, 12: 1415.
251. H. C. Huff, A. Vasan, A. Kaul, E. Tajkhorshid, and A. Das (2021)
Differential Interactions of Selected Phytocannabinoids with Human CYP2D6 Polymorphisms.
Biochemistry, 60, 2749–2760.
250. A. Rasouli, Y. Jamali, E. Tajkhorshid, O. Bavi, and H. Nejat Pishkenari (2021)
Mechanical properties of Ester- and Ether-DPhPC bilayers: a molecular dynamics study.
J. Mech. Behavior Biomedical Materials, 117, 104386.
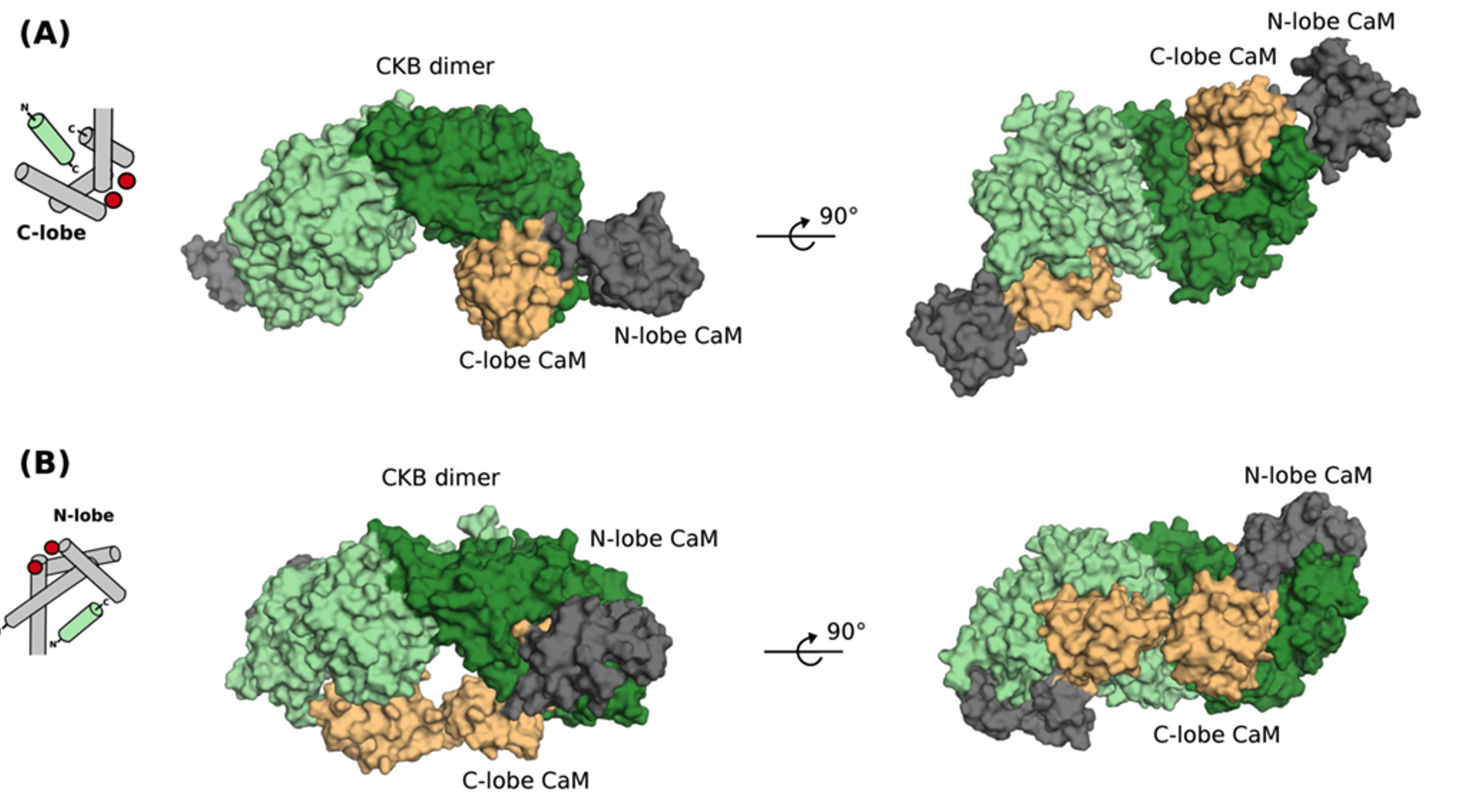
Calmodulin complexes with brain and muscle creatine kinase peptides.
Current Research in Structural Biology, 3: 121-132.
2020
Scalable molecular dynamics on CPU and GPU architectures with NAMD.
J. Chem. Phys., 153: 044130.
PMC7395834
Computational Dissection of Membrane Transport at a Microscopic Level.
Trends in Biochemical Sciences, 45: 202-216.
Twisting and tilting of a mechanosensitive molecular probe detects order in membranes.
Chemical Science, 11: 5637-49. (Cover Article)
PMC7433777
Microscopic Characterization of GRP1 PH Domain Interaction With Anionic Membranes.
Journal of Computational Chemistry, 41: 489-499. (Cover Article)
PMC7000246
Structural basis for the reaction cycle of DASS dicarboxylate transporters.
eLife, 9: e61350. PMC7553777
Associated eLife Insight Article: A. W. Duster and H. Lin (2020)
Riding elevators into and out of cells.
eLife, 9: e62925. PMC7553771
Boosting Free-Energy Perturbation Calculations with GPU-Accelerated NAMD.
J. Chem. Inf. Model., 60: 5301-5307.
PMC7686227
Probing Cholesterol Binding and Translocation in P-glycoprotein.
Biochimica et Biophysica Acta - Biomembranes, 1862: 183090. (Cover Article)
Membrane Interactions of Cy3/Cy5 Fluorophores and Their Effects on Membrane Protein Dynamics.
Biophysical Journal, 119: 24-34.
A CLC-ec1 mutant reveals global conformational change and suggests a unifying mechanism for the CLC Cl-/H+ transport cycle.
eLife, 9:e53479. PMC7253180
Commentary by Chris Miller: Q-cubed mutant cues clues to CLC antiport mechanism. J. General Physiology, 153: e202112868 (2021).
Menthol binding to the human α4β2 nicotinic acetylcholine receptor, facilitated by its strong partitioning in membrane.
J. Phys. Chem. B, 124: 1866-1880.
Cryo-EM structures of a lipid-sensitive pentameric ligand-gated ion channel embedded in a phosphatidylcholine-only bilayers.
PNAS, 117: 1788-1798.
PMC6983364
An Activity-based Sensing Approach for the Detection of Cyclooxygenase-2 in Live Cells.
Angewandte Chemie, 59: 3307-3314.
Membrane Surface Recognition by the ASAP1 PH Domain and Consequences for Interactions with the small GTPase Arf1.
Science Advances, 6: eabd1882.
PMC7527224
Aβ(1-42) tetramer and octamer structures reveal edge conductivity pores as a mechanism for membrane damage.
Nature Communications, 11: 3014.
PMC7296003
234. J. W. Smith, X. Jiang, H. An, A. M. Barclay, G. Licari,
E. Tajkhorshid, E. Moore, C. M. Rienstra, J. S. Moore and Q. Chen (2020)
Polymer-Peptide Conjugates Convert Amyloid into Protein Nanobundles through Fragmentation and Lateral Association.
ACS Applied Nano Materials, 2: 937-945.
(Cover Article)
Identifying mutation hotspots reveals pathogenetic mechanisms of KCNQ2 epileptic encephalopathy.
Scientific Reports, 10, 4756.
PMC7075958
Confronting pitfalls of AI-augmented molecular dynamics using statistical physics.
J. Chem. Phys., 153: 234118.
Editors' Choice
Cover Article
Featured Article
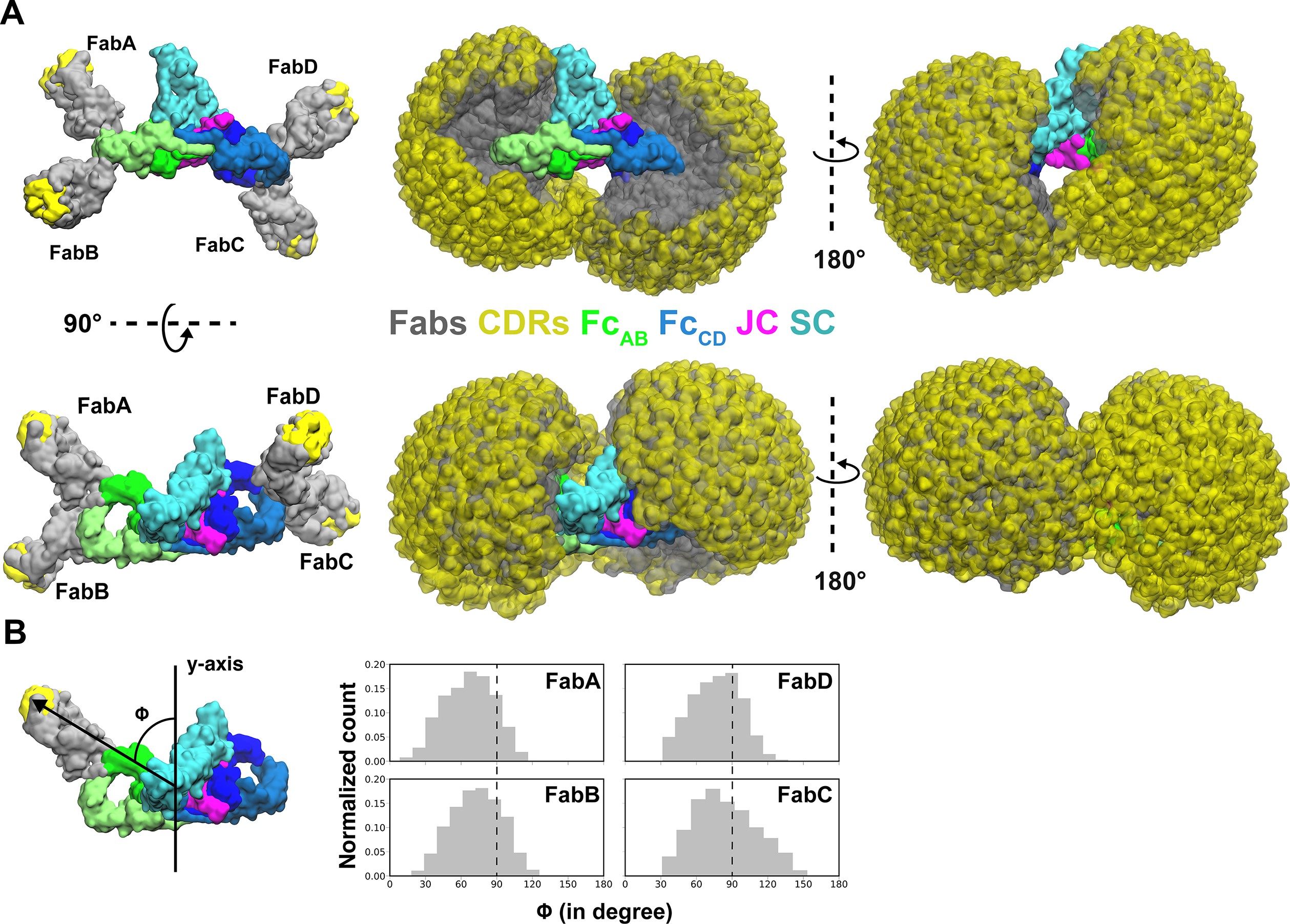
The structures of Secretory and dimeric Immunoglobulin A.
eLife, 9:e56098.
230. Y. T. Pang, A. Pavlova, E. Tajkhorshid, and J. C. Gumbart (2020)
Parameterization of a drug molecule with a halogen σ-hole particle
using ffTK: Implementation, testing and comparison.
J. Chem. Phys., 153: 164104.
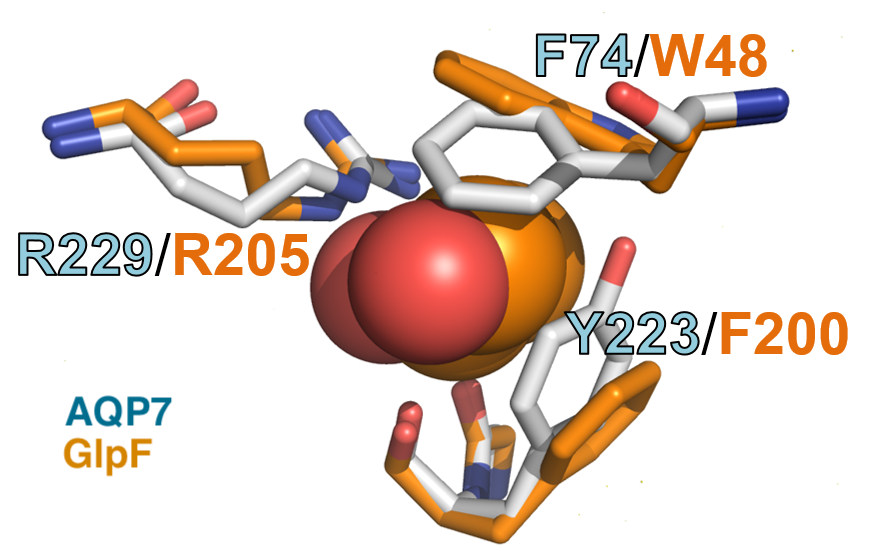
Aquaporin-7: A Dynamic Aquaglyceroporin with Greater Water and Glycerol Permeability than its Bacterial Homolog GlpF.
Frontiers in Physiology, 11, 728.
PMC7339978
Molecular Insights into the Loading and Dynamics of Doxorubicin on PEGylated Graphene Oxide Nanocarriers.
ACS Applied Bio Materials, 3, 1354−1363.
Aberrant expression of a non-muscle RBFOX2 isoform triggers cardiac conduction defects in myotonic dystrophy.
Developmental Cell, 52: 748-763.
Hydrogen-deuterium exchange mass spectrometry captures distinct dynamics upon substrate and inhibitor binding to a transporter.
Nature Communications, 11: 6162.
PMC7710758
225. M. Yang, J. Sun, D. F. Stowe, E. Tajkhorshid, W.-M. Kwok,
and A. K. S. Camara (2020)
Knockout of VDAC1 in H9c2 Cells Promotes Oxidative Stress-induced
Cell Apoptosis Through Decreased Mitochondrial Hexokinase II Binding
and Enhanced Glycolytic Stress.
Cell. Physiol. Biochem., 54: 853-874.
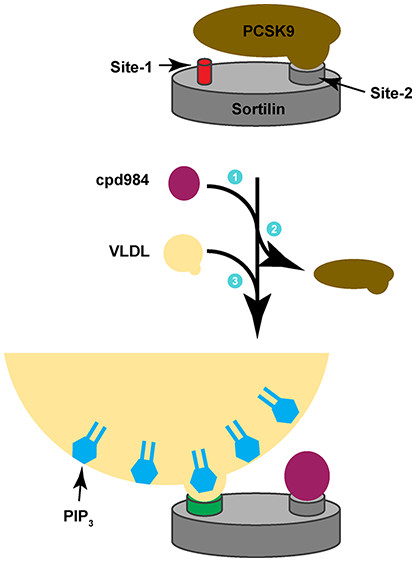
An Allosteric Binding Site on Sortilin Regulates the Trafficking of VLDL and PCSK9 Altering LDLR Expression Levels in Hepatocytes.
Biochemistry, 59: 4321-35.
Structural and functional diversity calls for a new classification of ABC transporters
FEBS Letters, 594: 3767-75.
2019
222. A. Singharoy*, C. Maffeo, K.H. Delgado-Magnero, D. J. K. Swainsbury, M.Sener,
U. Kleinekathofer, B. Isralewitz, I. Teo, D. Chandler, J. W. Vant, J. E. Stone,
J. Phillips, T. V. Pogorelov, M. I. Mallus, C. Chipot, Z. Luthey-Schulten,
P. Tieleman, C. N. Hunter, E. Tajkhorshid*, A. Aksimentiev*, and K. Schulten (2019)
Atoms to Phenotypes: Molecular Design Principles of Cellular Energy Metabolism.
Cell, 179: 1098-1111.
PMC7075482
221. M. P. Muller*, T. Jiang*, C. Sun, M. Lihan, S. Pant,
P. Mahinthichaichan, A. Trifan, and E. Tajkhorshid (2019)
Characterization of Lipid-Protein Interactions and Lipid-mediated Modulation of Membrane Protein Function Through Molecular Simulation.
TOP5 community's Favorite Review of 2019
Chemical Reviews, 119: 6068-6161.
220. K. Yu*, T. Jiang*, Y. Cui, E. Tajkhorshid*, and H. C. Hartzell* (2019)
A Network of Phosphatidylinositol 4,5-bisphosphate Binding Sites Regulate
Gating of the Ca2+-activated Cl- Channel ANO1 (TMEM16A).
Proceedings of the National Academy of Sciences USA, 116: 19952-19962.
219. C. Martens, M. Shekhar, A. M. Lau, E. Tajkhorshid*, and A. Politis* (2019)
Integrating hydrogen-deuterium exchange mass spectrometry with molecular dynamics simulations
to probe lipid-modulated conformational changes in membrane proteins.
Nature Protocols, 14: 3183-3204.
PMC7058097
218. J. A. Coleman , D. Yang , Z. Zhao , P.-C. Wen , C. Yoshioka , E. Tajkhorshid, and E. Gouaux (2019)
Serotonin transporter-ibogaine complexes illuminate mechanisms of inhibition and transport.
Nature, 569: 141-145.
PMC6925253
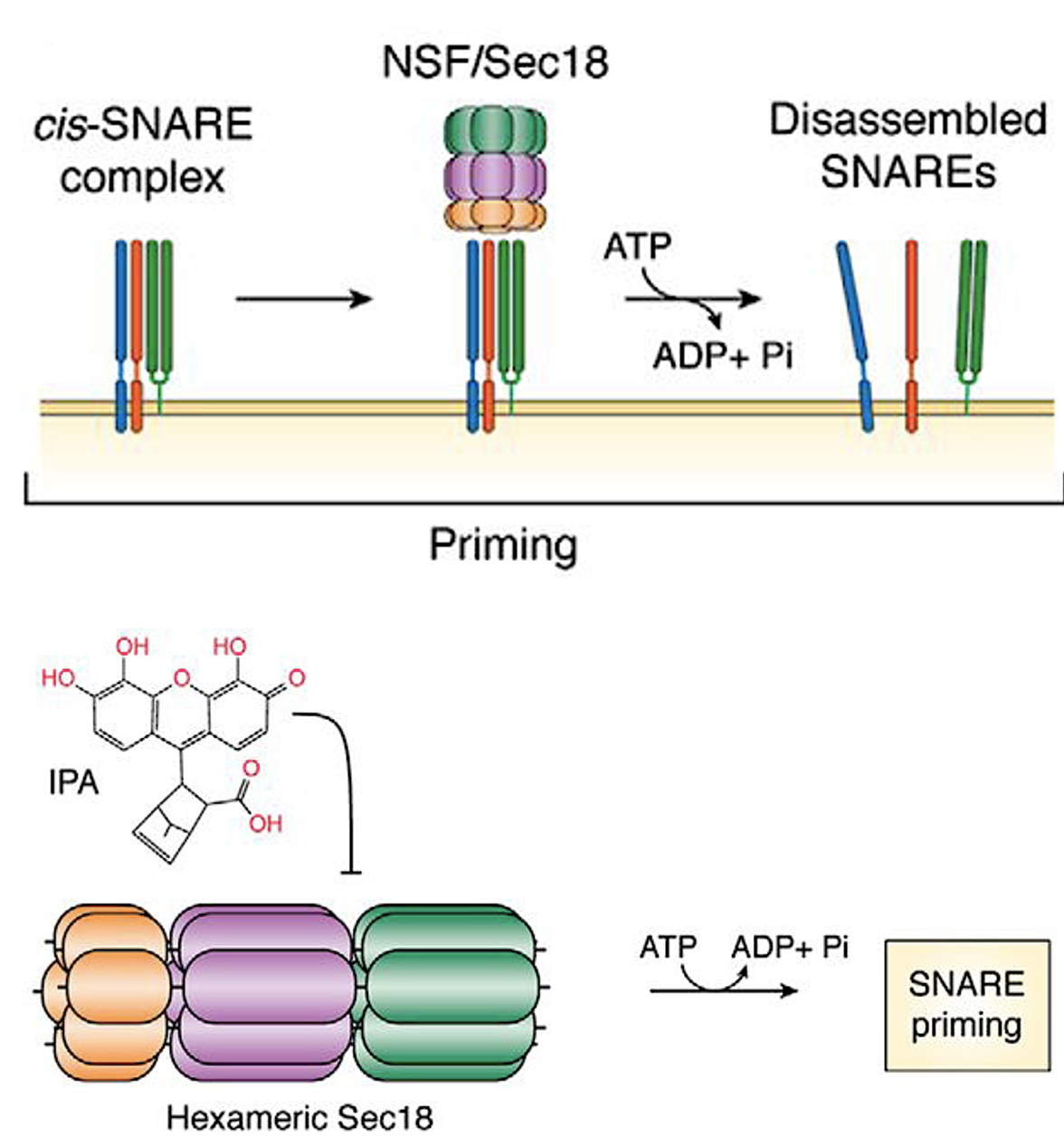
216. R. P. Sparks*, A. S. Arango*, M. L. Starr*, Z. L. Aboff, K. A. Harden,
J. L. Jenkins, W. C. Guida, E. Tajkhorshid, and R. A. Fratti (2019)
A Small Molecule Competitive Inhibitor of Phosphatidic Acid Binding
by Sec18 Blocks the SNARE Priming Stage of Vacuole Fusion.
J. Biol. Chem., 294: 17168-17185.
JBC EDITORS' PICK (top 2% in significance and importance)
PMC6873166
215. I. G. Denisov, Y. V. Grinkova, P. Nandigrami, M. S. Shekhar, E. Tajkhorshid, and S. G. Sligar (2019)
Allosteric Interactions in Human Cytochrome P450 CYP3A4: The Role of Phenylalanine 213.
Biochemistry, 58: 1411-1422.
214. X. Jiang, A. Halmes, G. Licari, J. Smith, Y. Song, E. Moore, Q. Chen*, E. Tajkhorshid*, C. Rienstra*, and J. Moore* (2019)
Multivalent Polymer-Peptide Conjugates-A General Platform for Inhibiting Amyloid Beta Peptide Aggregation.
ACS Macro Letters, 8, 1365-1371.
PMC7059649
213. S. K. Misra, Z. Wu, F. Ostadhossein, M. Ye, K. Boateng, K. Schulten, E. Tajkhorshid, and D. Pan (2019)
Pro-nifuroxazide Self-Assembly Leads to Triggerable Nanomedicine for Anti-Cancer Therapy.
ACS Applied Materials & Interfaces, 11, 18074-18089.
212. A. Aster, G. Licari, F. Zinna, E. Brun, T. Kumpulainen, E. Tajkhorshid, J. Lacour and E. Vauthey (2019)
Tuning symmetry breaking charge separation in perylene bichromophores by conformational control.
Chemical Science, 10, 10629-10639.
211. M. L. Starr*, R. P. Sparks*, A. Arango*, L. R. Hurst, Z. Zhao, M. Lihan,
J. L. Jenkins, E. Tajkhorshid, and R. A. Fratti (2019)
Phosphatidic acid induces conformational changes in Sec18 protomers that prevent SNARE priming.
J. Biol. Chem., 294: 3100-3116.
Structural Insights into the Lipid A Transport Pathway in MsbA.
Structure, 27, 1114-1123.e3.
A Chalcogen-Bonding Cascade Switch for Planarizable Push-Pull Probes.
Angewandte Chemie, 58, 15752-15756.
PMC7035594
208. K. Terekhova, S. Pokutta, Y. S. Kee, J. Li, E. Tajkhorshid,
G. Fuller, A. R. Dunn, and W. I. Weis (2019)
Binding partner- and force-promoted changes in αE-catenin conformation probed by native cysteine labeling.
Scientific Reports, 9, 15375.
PMC6814714
Federating Structural Models and Data: Outcomes from a Workshop on Archiving Integrative Structures.
Structure , 27: 1745-1759.
2018

206. J. V. Vermaas, S. Rempe, and E. Tajkhorshid (2018)
Electrostatic Lock in the Transport Cycle of the Multi-Drug Resistance Transporter EmrE.
PNAS, 115: E7502-E7511.
Commentary by Jana Shen: Zooming in on a small multidrug transporter reveals details of asymmetric protonation.
PNAS, 115: 8060-8062.
Probing key elements of teixobactin-lipid II interactions in membrane.
Chemical Science, 9: 6997-7008. (Open Access)
204. C. Sun, S. Benlekbir, P. Venkatakrishnan, Y. Wang, S. Hong, J. Hosler, E. Tajkhorshid*, J. L. Rubinstein*, and R. B. Gennis* (2018)
Structure of the Alternative Complex III in a supercomplex with cytochrome oxidase.
Nature, 557: 123-126
Microscopic View of Lipids and Their Diverse Biological Functions.
Current Opinion in Structural Biology, 51: 177-186.
Direct protein-lipid interactions shape the conformational landscape of secondary transporters.
Nature Communications, 9: 4151.
201. D. T. Infield, K. Matulef, J. D. Galpin, K. Lam, E. Tajkhorshid, C. A. Ahern, and F. I. Valiyaveetil (2018)
Main-chain mutagenesis reveals intrahelical coupling in an ion channel voltage-sensor.
Nature Communications, 9: 5055.
Selective permeability of carboxysomes shell pores to anionic molecules.
J. Phys. Chem. B, 122: 9110-9118.
Cytochrome aa3 oxygen reductase utilizes the tunnel observed in the crystal structures to deliver O2 for catalysis.
Biochemistry, 57: 2150-2161.
198. Y. Wang*, M. Shekhar*, D, Thifault, C. J. Williams, R. Mcgreevy, J. Richardson, A. Singharoy*, and E. Tajkhorshid* (2018)
Constructing Atomic Structural Models into Cryo-EM densities using Molecular Dynamics - Pros and Cons.
Journal of Structural Biology, 204: 319-328.
PyContact: Rapid, Customizable and Visual Analysis of Non-Covalent Interactions in MD Simulations.
Biophysical Journal, 114: 577-583. (Cover Article)
196. B. Henderson, S. Grant, B. Chu, R. Shahoei, S. Huard, S. Saladi,
E. Tajkhorshid, D. Dougherty, and H. Lester (2018)
Menthol stereoisomers exhibit different effects on α4β2
nAChR upregulation and dopamine neuron spontaneous firing.
eNeuro, ENEURO.0465-18.2018.
194. P. Mahinthichaichan, R. B. Gennis*, and E. Tajkhorshid* (2018) Bacterial denitrifying nitric oxide reductases and aerobic respiratory terminal oxidases use similar delivery pathways for their molecular substrates. BBA - Bioenergetics, 1859: 712-724.
193. I. G. Denisov, J. L. Baylon, Y. V. Grinkova, E. Tajkhorshid, and S. G. Sligar (2018) Drug-drug interactions between atorvastatin and dronedarone mediated by monomeric CYP3A4. Biochemistry, 57: 805-816.
192. S. Barrick, J. Li, X. Kong, A. Ray, E. Tajkhorshid, and D. Leckband (2018) Salt Bridges Gate Alpha-Catenin Activation at Intercellular Junctions. Mol. Biol. Cell, 29: 111-122.
191. A. Chakravarti, K. Selvadurai, R. Shahoei, H. Lee, S. Fatma, E. Tajkhorshid, and R. Huang (2018) Reconstitution and substrate specificity of the antiviral radical SAM enzyme viperin. J. Biol. Chem., 293: 14122-14133..
189. T. E. Speltz*, C.G. Mayne*, S.E. Fanning, Z. Siddiqui, E. Tajkhorshid, G.L. Greene, and T.W. Moore (2018) A "Cross-Stitched" Peptide with Improved Helicity and Proteolytic Stability. Organic & Biomolecular Chemistry, 16: 3702-3706.
188. M. Hallock, A. Greenwood, Y. Wang, J. Morrissey, E. Tajkhorshid, C. Rienstra, and T. V. Pogorelov (2018) Calcium-induced lipid nanocluster structures: sculpturing of the plasma membrane. Biochemistry, 57: 6897-6905.
187. D. Kerr, G. T. Tietjen, Z. Gong, E. Tajkhorshid, E. Adams, and K.-Y. C. Lee (2018) Sensitivity of Peripheral Membrane Proteins to the Membrane Context: A Case Study of Phosphatidylserine and the TIM Proteins BBA - Biomembranes, 1860: 2126-2133.
186. S. W. Fanning, R. Jeselsohn, V. Dharmarajan, C. G. Mayne, M. Karimi, G. Buchwalter, R. Houtman, W. Toy, C. E. Fowler, M. Lainé, K. E Carlson, T. A. Martin, J. Nowak, J. Nwachukwu, D. J. Hosfield, S. Chandarlapaty, E. Tajkhorshid, K. W. Nettles, P. R. Griffin, Y. Shen, J. A. Katzenellenbogen, M. Brown, and G. L. Greene (2018) The SERM/SERD Bazedoxifene Disrupts ESR1 Helix 12 to Overcome Acquired Hormone Resistance in Breast Cancer Cells. eLife, 2018;7: e37161. (Accompanied by a Related Insight article)
2017
184. S. Chen, Y. Zhao, Y. Wang, M. Shekhar, E. Tajkhorshid, and E. Gouaux (2017) Activation and desensitization mechanism of AMPA receptor-TARP complex by cryo-EM. Cell, 170(6): 1234-1246.
182. F. Zeng, Y. Chen, J. Remis, M. S. Shekhar, J. C. Phillips, E. Tajkhorshid, and H. Jin (2017) Structural Basis of Co-translational Quality Control by ArfA and RF2 Binding to Ribosome. Nature, 541, 554-557.

178. J. V. Vermaas and E. Tajkhorshid (2017) Differential Membrane Binding Mechanics of Synaptotagmin Isoforms Observed at Atomic Detail. Biochemistry, 56: 281-293.

174. W. R. Arnold, J. L. Baylon, E. Tajkhorshid*, and A. Das* (2017) Arachidonic Acid Metabolism by Human Cardiovascular CYP2J2 is Modulated by Doxorubicin. Biochemistry, 56: 6700-6712.
172. X. Yu, G. Yang, C. Yan, J. L. Baylon, J. Jiang, H. Fan, G. Lu, K. Hasegawa, H. Okumura, T. Wang, E. Tajkhorshid, S. Li, and N. Yan (2017) Dimeric structure of the uracil:proton symporter UraA provides mechanistic insights into the SLC4/23/26 transporters. Cell Research, 27: 1020-1033.
171. P. S. Padayatti, J. H. Leung, P. Mahinthichaichan, E. Tajkhorshid, A. Ishchenko, V. Cherezov, M. Soltis, B. J. Jackson, C. D. Stout, R. B. Gennis, and Q. Zhang (2017) Critical role of water molecules in proton translocation by the membrane-bound transhydrogenase. Structure, 25(7): 1111-1119.e3.
170. A. K. S. Camara, Y. Zhou, P.-C. Wen, E. Tajkhorshid and W.-M. Kwok (2017) Mitochondrial VDAC1: A Key Gatekeeper as Potential Therapeutic Target. Frontiers in Physiology, 8: 640.
2016
N. Trebesch, J. V. Vermaas, E. Tajkhorshid (2016) Computational Characterization of Molecular Mechanisms of Membrane Transporter Function. In Editor: Carmen Domene, "Computational Biophysics of Membrane Proteins", Chapter 7: pp. 197-236. Royal Society of Chemistry, Cambridge, UK.
T. Jiang, W. Han, M. Maduke, and E. Tajkhorshid (2016) Molecular Basis for Differential Anion Binding and Proton Coupling in the Cl-/H+ Exchanger ClC-ec1. Journal of the American Chemical Society, 138: 3066-3075.
C. M. Khantwal, S. J. Abraham, W. Han, T. Jiang, T. S. Chavan, R. C. Cheng, S. M. Elvington, C. W. Liu, I. I. Mathews, E. Tajkhorshid*, and M. Maduke* (2016) Revealing an outward-facing open conformational state in a CLC Cl/H exchange transporter. eLife, 5:e11189.
J. Vermaas, N. Trebesch, C. G. Mayne, S. Thangapandian, M. Shekhar, P. Mahinthichaichan, J. L. Baylon, T. Jiang, Y. Wang, M. P. Muller, E. Shinn, Z. Zhao, P.-C. Wen, and E. Tajkhorshid (2016) Microscopic Characterization of Membrane Transport Function by In Silico Modeling and Simulation. In Gregory A. Voth, editor: Methods in Enzymology, Vol 578: Computational Approaches for Studying Enzyme Mechanism Part B, MIE, UK: Academic Press, 2016, pp. 373-428.
P. Mahinthichaichan, R. Gennis*, and E. Tajkhorshid* (2016) All the O2 consumed by Thermus thermophilus cytochrome ba3 is delivered to the active site through a long, open hydrophobic tunnel with entrances within the lipid bilayer. Biochemistry, 55(8): 1265-1278..
J. L. Baylon, J. V. Vermaas, M. P. Muller, M. J. Arcario, T. V. Pogorelov, and E. Tajkhorshid (2016) Atomic-Level Description of Protein--Lipid Interactions Using an Accelerated Membrane Model. Biochimica et Biophysica Acta - Biomembranes, 1858: 1573-1583.
K. K. Skeby, O. J. Andersen, T. V. Pogorelov, and E. Tajkhorshid*, and B. Schiøtt* (2016) Conformational Dynamics of the Human Islet Amyloid Polypeptide in a Membrane Environment: Toward the Aggregation Prone Form. Biochemistry, 55: 2031-2042.
W. R. Arnold, J. L. Baylon, E. Tajkhorshid*, and A. Das* (2016) Asymmetric binding and metabolism of polyunsaturated fatty acids (PUFAs) by CYP2J2 epoxygenase. Biochemistry, 55: 6969-6980.
J. Vermaas, D. Hardy, J. Stone, E. Tajkhorshid*, and A. Kohlmeyer* (2016) TopoGromacs: Automated Topology Conversion from CHARMM to Gromacs within VMD. Journal of Chemical Information and Modeling, 56: 1112-1116.
P. Hosseinzadeh, E. Mirts, T. Pfister, Y-G. Gao, C. G. Mayne, H. Robinson, E. Tajkhorshid, and Y. Lu (2016) Enhancing Mn(II)-binding and manganese peroxidase activity in a designed cytochrome c peroxidase through fine-tuning secondary sphere interactions. Biochemistry, 55(10): 1494-1502.
S. W. Fanning*, C. G. Mayne*, V. Dharmarajan*, K. E. Carlson, T. A. Martin, S. J. Novick, W. Toy, B. Green, S. Panchamukhi, B. S. Katzenellenbogen, E. Tajkhorshid, P. R. Griffin, Y. Shen, S. Chandarlapaty, J. A. Katzenellenbogen, and G. L. Greene (2016) Estrogen receptor alpha somatic mutations Y537S and D538G confer breast cancer endocrine resistance by stabilizing the activating function-2 binding conformation. eLife, 10.7554/eLife.12792. (Accompanied by a "Related Insight" Article)
C. Sun, A. T. Taguchi, J. V. Vermaas, N. J. Beal, P. J. O'Malley*, E. Tajkhorshid*, R. B. Gennis*, and S. A. Dikanov* (2016) Q-band Electron-Nuclear Double Resonance Reveals Out-of-Plane Hydrogen Bonds Stabilize an Anionic Ubisemiquinone in Cytochrome bo3 from Escherichia coli. Biochemistry, 55: 5714-5725.
2015
J. Li, P.-C. Wen, M. Moradi, and E. Tajkhorshid (2015) Computational Characterization of Structural Dynamics Underlying Function in Active Membrane Transporters Current Opinion in Structural Biology, 31: 96-105. PMC4476910
J. Li, J. Newhall, N. Ishiyama, C. Gottardi, M. Ikura, D. E. Leckband*, and E. Tajkhorshid* (2015) Structural Determinants of the Mechanical Stability of Alpha Catenin. Journal of Biological Chemistry, 290: 18890-18903.
J. L. Baylon and E. Tajkhorshid (2015) Capturing Spontaneous Membrane Insertion of the Influenza Virus Hemagglutinin Fusion Peptide. Journal of Physical Chemistry B, 119(25): 7882-7893.
J. V. Vermaas, J. L. Baylon, M. J. Arcario, M. P. Muller, Zh. Wu, T. V. Pogorelov, and E. Tajkhorshid (2015) Efficient Exploration of Membrane-Associated Phenomena at Atomic Resolution. Journal of Membrane Biology, 248: 563-582. PMC4490090
J. V. Vermaas, A. T. Taguchi, S. A. Dikanov, C. A. Wraight, and E. Tajkhorshid (2015) Redox Potential Tuning Through Differential Quninone Binding in the Photosynthetic Reaction Center of Rhodobacter sphaeroides Biochemistry, 54: 2104-2116.
I. G. Denisov, Y. V. Grinkova, J. L. Baylon, E. Tajkhorshid, and S. G. Sligar (2015) Mechanism of Drug-Drug Interactions Mediated by Human Cytochrome P450 CYP3A4 Biochemistry, 54: 2227-2239.
D. R. McDougle*, J. L. Baylon*, D. D. Meling, A. Kambalyal, Y. V. Grinkova, J. Hammernik, E. Tajkhorshid*, and A. Das* (2015) Incorporation of Charged Residues in the CYP2J2 F-G Loop Disrupts CYP2J2-Lipid Bilayer Interactions. Biochimica et Biophysica Acta - Biomembranes, 1848: 2460-2470.
J. Madsen, Y. Z. Ohkubo, G. Peters, J. H. Faber, E. Tajkhorshid*, and O. H. Olsen* (2015) Membrane interaction of the factor VIIIa discoidin domains in atomistic details. Biochemistry, 54: 6123-6131.
Y. Qi, X. Cheng, J. Lee, J. Vermaas, T. V. Pogorelov, E. Tajkhorshid, S. Park, J. B. Klauda, and W. Im (2015) CHARMM-GUI HMMM Builder for Membrane Simulations with the Highly Mobile Membrane-Mimetic Model. Biophysical Journal, 109: 2012-22.
2014
M. J. Arcario, C. G. Mayne, and E. Tajkhorshid (2014) Atomistic models of general anesthetics for use in in silico biological studies. Journal of Physical Chemistry B, 118: 12075-12086.
W. Han, R. Chang, M. Maduke, and E. Tajkhorshid (2014) Water Access Points and Hydration Pathways in the ClC-ec1 H+/Cl- Transporters. Proceedings of the National Academy of Sciences USA, 111: 1819-1824. (Cover Article)
R. E. Hulse, J. Sachleben, P.-C. Wen, M. Moradi, E. Tajkhorshid, and E. Perozo (2014) Conformational dynamics at the inner gate of KcsA during activation. Biochemistry (Rapid Report), 53: 2557-2559. (live molecular demo sessions of KcsA channel)
S. Mishra, B. Verhalen, R. A. Stein, P.-C. Wen, E. Tajkhorshid, and H. S. Mchaourab (2014) Conformational Dynamics of the Nucleotide Binding Domains and the Power Stroke of a Heterodimeric ABC Transporter. eLife, 3: e02740
M. Moradi and E. Tajkhorshid (2014) Computational recipe for efficient description of large-scale conformational changes in biomolecular systems. Journal of Chemical Theory and Computation, 10: 2866-2880. PMCID: PMC4089915
A. Barati Farimani, N. Aluru, and E. Tajkhorshid (2014) Thermodynamic Insight into Spontaneous Hydration and Rapid Water Permeation in Aquaporins. Applied Physics Letters, 105, 083702.
J. V. Vermaas and E. Tajkhorshid (2014) Conformational Heterogeneity of alpha-synuclein in Membrane. Biophysica Biochemica Acta - Biomembranes, 1838: 3107-3117.
M. J. Arcario and E. Tajkhorshid (2014)
Membrane-Induced Structural Rearrangement of Talin Subdomains Providing a Molecular Mechanism for Inside-Out Activation of Integrin.
Biophysical Journal, 107: 2059-2069.
Animation highlighted on the Biophysical Journal page
A. E. Blanchard*, M. J. Arcario*, K. Schulten, and E. Tajkhorshid (2014)
A Highly Tilted Membrane Configuration For Pre-Fusion State of Synaptobrevin.
Biophysical Journal, 107: 2112-2121.
* These authors contributed equally to the paper.
Y. Ahn, P. Mahinthichaichan, H. J. Lee, H. Ouyang, D. Kaluka, S.-R. Yeh, D. Arjona, D. L. Rousseau, E. Tajkhorshid, P. Ädelroth, and R. B. Gennis (2014) Conformational coupling between the active site and residues within the KC-channel of the Vibrio cholerae cbb3-type oxygen reductase. Proceedings of the National Academy of Sciences USA, 111: E4419-E4428.
M.-R. Kalani and E. Tajkhorshid (2014) Molecular Dynamics: The Computational Molecular Microscope. Razavi International Journal of Medicine, 2(3): e20117.
S. N. Smith, Y. Wang, J. L. Baylon, N. Singh, B. Baker, E. Tajkhorshid, and D. M. Kranz (2014) Changing the peptide specificity of a human T cell receptor by directed evolution. Nature Communication, 5:5223.
G. Enkavi, J. Li, P.-C. Wen, S. Thangapandian, M. Moradi, T. Jiang, W. Han, and E. Tajkhorshid (2014) A Microscopic View of the Mechanisms of Active Transport Across the Cellular Membrane. Annual Reports in Computational Chemistry, 10:77-125.
2013
J. Li, S. A. Shaikh, G. Enkavi, P.-C. Wen, Z. Huang and E. Tajkhorshid (2013) Transient Formation of Water-conducting States in Membrane Transporters. Proceedings of the National Academy of Sciences USA, 110: 7696-7701. PMCID: PMC3651479
U. K. Eriksson, G. Fischer, R. Friemann, G. Enkavi, E. Tajkhorshid*, and R. Neutze* (2013) Sub-Angstrom resolution x-ray structure details aquaporin-water interactions. Science, 340: 1346-1349.
S. A. Shaikh, J. Li, G. Enkavi, P.-C. Wen, Z. Huang, and E. Tajkhorshid (2013) Visualizing Functional Motions of Membrane Transporters with Molecular Dynamics Simulations. Biochemistry (Current Topic), 52: 569-587. PMCID: PMC3560430
J. L. Baylon, I. Lenov, S. G. Sligar, and E. Tajkhorshid (2013) Characterizing the Membrane-Bound State of Cytochrome P450 3A4: Structure, Depth of Insertion and Orientation. Journal of the American Chemical Society, 135: 8542-8551.
M. Moradi and E. Tajkhorshid (2013) Driven Metadynamics: Reconstructing Equilibrium Free Energies From Driven Adaptive-Bias Simulations. Journal of Physical Chemistry Letters, 4: 1882-1887.
P.-C. Wen, B. Verhalen, S. Wilkens, H. Mchaourab, and E. Tajkhorshid (2013) On the Origin of Large Flexibility of P-glycoprotein in the Inward-Facing State. Journal of Biological Chemistry, 288: 19211-19220.
M. R. Kalani, A. Moradi, M. Moradi, and E. Tajkhorshid (2013) Characterizing a Histidine Switch Controlling pH-Dependent Conformational Changes of the Influenza Virus Hemagglutinin. Biophysical Journal, 105: 993-1003.
G. Enkavi, J. Li, P. Mahinthichaichan, P.-C. Wen, Z. Huang, S. A. Shaikh, and E. Tajkhorshid (2013) Simulation Studies of the Mechanism of Membrane Transporters. In Editors: Luca Monticelli and Emppu Salonen, "Biomolecular Simulations - Methods and Protocols", Humana Press. Methods in Molecular Biology, Vol. 924, Part 2, 361-405.
R. R. Geyer, R. Musa-Aziz, G. Enkavi, P. Mahinthichaichan, E. Tajkhorshid, and W. Boron (2013) Movement of NH3 through the Human Urea Transporter B (UT-B): A New Gas Channel. American Journal of Physiology - Renal Physiology, 304: F1447-F1457.
2012
E. J. Levin, Y. Cao, G. Enkavi, M. Quick, Y Pan, E. Tajkhorshid*, and M. Zhou* (2012) Structure and permeation mechanism of a mammalian urea transporter. Proceedings of the National Academy of Sciences USA, 109: 11194-11199.
N. Noinaj, N. C. Easley, M. Oke, N. Mizuno, J. C. Gumbart, E. Boura, A. N. Steere, O. Zak, P. Aisen, E. Tajkhorshid, R. W. Evans, A. R. Gorringe, A. B. Mason, A. C. Steven, and S. K. Buchanan (2012) Structural basis for iron piracy by pathogenic Neisseria. Nature, 483: 53-58
J. Li and E. Tajkhorshid (2012) A Gate-Free Pathway for Substrate Release from the Inward-Facing State of the Na+-Galactose Transporter. Biochimica et Biophysica Acta (BBA) - Biomembranes, 1818: 263-271. NIHMSID: 329267
M. Musgaard, L. Thøgersen, B. Schiøtt, and E. Tajkhorshid (2012) Tracing Cytoplasmic Ca2+ Ion and Water Access Points in the Ca2+-ATPase. Biophysical Journal, 102: 268-277.
T. J. Barnard, J. C. Gumbart, J. H. Peterson, N. Noinaj, N. C. Easley, N. Dautin, A. J. Kuszak, E. Tajkhorshid, H. D. Bernstein, and S. K. Buchanan (2012) Molecular Basis for Activation of a Catalytic Asparagine Residue in a Self-Cleaving Bacterial Autotransporter. Journal of Molecular Biology, 415: 128-142.
F. Khalili-Araghi, E. Tajkhorshid, B. Roux, and K. Schulten (2012) Molecular dynamics investigation of the omega current in the Kv1.2 voltage sensor domains. Biophysical Journal, 102: 258-267.
J. H. Morrissey, E. Tajkhorshid, S. G. Sligar, and C. M. Rienstra (2012) Tissue Factor/Factor VIIa Complex: Role of the Membrane Surface. Thrombosis Research, 129: S8-S10.
2011
P.-C. Wen and E. Tajkhorshid (2011) Conformational Coupling of the Nucleotide-Binding and the Transmembrane Domains in ABC Transporters. Biophysical Journal, 101: 680-690.
M. J. Arcario, Y. Z. Ohkubo, and E. Tajkhorshid (2011) Capturing Spontaneous Partitioning of Peripheral Proteins using a Biphasic Membrane-Mimetic Model. Journal of Physical Chemistry B, 115: 7029-7037. PMCID: PMC3102442.
J. Boettcher, R. Davis-Harrison, M. Clay, A. Nieuwkoop, Y. Z. Ohkubo, E. Tajkhorshid, J. H. Morrissey, and C. Rienstra (2011) Atomic View of Calcium-Induced Clustering of Phosphatidylserine in Mixed Lipid Bilayers. Biochemistry, 50: 2264-2273.
Z. Huang, S. A. Shaikh, P.-C. Wen, G. Enkavi, J. Li, and E. Tajkhorshid (2011) Membrane Transporters - Molecular Machines Coupling Cellular Energy to Vectorial Transport Across the Membrane. In Editor: Benoit Roux, "Molecular Machines", Chapter 9: pp. 151-182, World Scientific Publishing, Singapore.
J. H. Morrissey, E. Tajkhorshid, and C. M. Rienstra (2011) Nanoscale studies of protein-membrane interactions in blood clotting. Journal of Thrombosis and Haemostasis, 9 (Suppl. 1): 162-167.
2010
Z. Huang and E. Tajkhorshid (2010) Identification of the Third Na+ Site and the Sequence of Extracellular Binding Events in the Glutamate Transporter. Biophysical Journal, 99: 1416-1425.
S. Shaikh and E. Tajkhorshid (2010) Modeling and dynamics of the inward-facing state of a Na+/Cl- dependent neurotransmitter transporter homologue. PLoS Computational Biology, 6(8): e1000905.
Y. Z. Ohkubo, J. H. Morrissey, and E. Tajkhorshid (2010) Dynamical view of human tissue factor, factor VIIa, and their complex in solution and on the surface of anionic membranes. Journal of Thrombosis and Hemostasis, 8: 1044-1053. PMCID: PMC2890040.
Y. Wang, S. A. Shaikh, and E. Tajkhorshid (2010) Exploring transmembrane diffusion pathways with molecular dynamics. Physiology, 25: 142-145.
G. Enkavi and E. Tajkhorshid (2010) Simulation of spontaneous substrate binding revealing the binding pathway and mechanism and initial conformational response of GlpT. Biochemistry, 49: 1105-1114. PMCID: PMC2829668.
Y. Wang and E. Tajkhorshid (2010) Nitric oxide conduction by the brain aquaporin AQP4. PROTEINS, Structure, Function, and Bioinformatics, 78(3): 661-670. PMCID: PMC2805761.
J. H. Morrissey, R. L. Davis-Harrison, N. Tavoosi, K. Ke, V. Pureza, J. M. Boettcher, M. C. Clay, C. M. Rienstra, Y. Z. Ohkubo, T. V. Pogorelov, and E. Tajkhorshid (2010) Protein-Phospholipid interactions in blood clotting. Thrombosis Research, 125 (Suppl. 1): S23-S25. PMCID: PMC2838931.
O. Kokhan, C. A. Wraight, and E. Tajkhorshid (2010) The Binding Interface of Cytochrome C and Cytochrome C1 in the BC1 Complex: Rationalizing the Role of Key Residues. Biophysical Journal, 99: 2647-2656.
J. Diao, A. J. Maniotis, R. Follberg, and E. Tajkhorshid (2010) Interplay of mechanical and binding properties of fibronectin type I. Theoretical Chemistry Accounts, 125: 397-405. PMCID: PMC2932639.
F. Khalili-Araghi, V. Jogini, V. Yarov-Yarovoy, E. Tajkhorshid, B. Roux, and K. Schulten (2010) Calculation of the gating charge for the Kv1.2 voltage-activated potassium channel. Biophysical Journal, 98: 2189-2198.
P.-C. Wen, Z. Huang, G. Enkavi, Y. Wang, J. C. Gumbart, and E. Tajkhorshid (2010) Molecular mechanisms of active transport across the cellular membrane. In, Editors: Mark Sansom and Philip Biggin, "Molecular Simulations and Biomembranes: From Biophysics to Function", pp. 248-286. Royal Society of Chemistry, Cambridge, UK.
S. Shaikh, P.-C. Wen, G. Enkavi, Z. Huang, and E. Tajkhorshid (2010) Capturing Functional Motions of Membrane Channels and Transporters with Molecular Dynamics Simulation. Journal of Computational and Theoretical Nanosciences, 7: 2481-2500.
J. Feng, E. Lucchinetti, G. Enkavi, Y. Wang, P. Gehrig, B. Roschitzki, M. C. Schaub, E. Tajkhorshid, K. Zaugg, and M. Zaugg (2010) Tyrosine phosphorylation by Src within the cavity of the adenine nucleotide translocase 1 regulates ADP/ATP exchange in mitochondria. American Journal of Physiology - Cell Physiology, 298: 740-748.
X.-D. Yang, E. Tajkhorshid, and L.-F. Chen (2010) Functional Interplay between Acetylation and Methylation of the RelA Subunit of NF-κB. Molecular and Cellular Biology, 30: 2170-2180.
H.-C. Siebert, M. Burg-Roderfeld, Th. Eckert, S. Stötzel, U. Kirch, T. Diercks, M. J. Humphries, M. Frank, R. Wechselberger, E. Tajkhorshid, and S. Oesser (2010) Interaction of the α2A domain of integrin with small collagen fragments. Protein and Cell, 1: 393-405.
2009
J. Li and E. Tajkhorshid (2009) Ion-releasing State of A Secondary Membrane Transporter. Biophysical Journal, 97: L29-L31.
Ch. Law, G. Enkavi, D.-N. Wang, and E. Tajkhorshid (2009)
Structural basis of substrate selectivity in the glycerol-3-phosphate:phosphate antiporter GlpT.
Biophysical Journal, 97: 1346-1353.
PMCID: PMC2749764.
J. C. Gumbart, M. C. Wiener, and E. Tajkhorshid (2009)
Coupling of calcium and substrate binding through loop alignment in the outer membrane transporter BtuB.
Journal of Molecular Biology, 393: 1129-1142.
PMCID: PMC2775145.
F. Khalili-Araghi, J. C. Gumbart, P.-C. Wen, M. Sotomayor, E. Tajkhorshid, and K. Schulten (2009) Molecular dynamics simulations of membrane channels and transporters. Current Opinion in Structural Biology, 19: 128-137.
M. Nyblom, A. Frick, Y. Wang, M. Ekvall, K. Hallgren, K. Hedfalk, R. Neutze, E. Tajkhorshid, and S. Tornroth-Horsefield (2009) Structural and Functional Analysis of SoPIP2;1 Mutants Adds Insight into Plant Aquaporin Gating. Journal of Molecular Biology, 387: 653-668.
S. Hayashi, E. Tajkhorshid, and K. Schulten (2009) Photochemical Reaction Dynamics of the Primary Event of Vision Studied by Means of a Hybrid Molecular Simulation. Biophysical Journal, 96: 403-416.
J. Dittmer, L. Thogersen, J. Underhaug, K. Bertelsen, T. Vosegaard, J. M. Pedersen, B. Schiott, E. Tajkhorshid, T. Skrydstrup, and N. C. Nielsen (2009) Incorporation of Antimicrobial Peptides into Membranes: A Combined Liquid-State NMR and Molecular Dynamics Study of Alamethicin in DMPC/DHPC Bicelles. Journal of Physical Chemistry B, 113: 6928-6937.
J. H. Morrissey, V. Pureza, R. L. Davis-Harrison, S. G. Sligar, C. M. Rienstra, A. Z. Kijak, Y. Z. Ohkubo, and E. Tajkhorshid (2009) Protein-membrane interactions: Blood clotting on nanoscale bilayers. Journal of Thrombosis and Haemostasis, 7 (Suppl. 1): 169-172.
K. Bertelsen, B. Paaske, L. Thogersen, E. Tajkhorshid, B. Schiott, T. Skrydstrup, N. Chr. Nielsen, and Th. Vosegaard (2009) Residue-Specific Information about the Dynamics of Antimicrobial Peptides from 1H-15N and 2H Solid-State NMR Spectroscopy. Journal of the American Chemical Society, 131: 18335-18342.
2008
Y. Wang and E. Tajkhorshid (2008) Electrostatic funneling of substrate in mitochondrial membrane carriers. Proceedings of the National Academy of Sciences USA, 105: 9598-9603.
Z. Huang and E. Tajkhorshid (2008) Dynamics of the Extracellular Gate and Ion-Substrate Coupling in the Glutamate Transporter. Biophysical Journal, 95: 2292-2300.
Y. Zenmei Ohkubo and E. Tajkhorshid (2008) Distinct Structural and Adhesive Roles of Ca2+ in Membrane Binding of Blood Coagulation Factors. Structure, 16: 72-81.
L. Celik, B. Schiott, and E. Tajkhorshid (2008) Substrate binding and formation of an occluded state in the leucine transporter. Biophysical Journal, 94: 1600-1612.
Y. Wang, Y. Zenmei Ohkubo and E. Tajkhorshid (2008) Gas conduction of lipid bilayers and membrane channels. In Scott Feller, Editor, Computational Modeling of Membrane Bilayers. Elsevier, Oxford, UK. Current Topics in Membranes, 60: 343-367.
J. Diao and E. Tajkhorshid (2008) Indirect role of Ca2+ in the assembly of the extracellular matrix proteins. Biophysical Journal, 95: 120-127.
J. H. Morrissey, V. Pureza, R. L. Davis-Harrison, S. G. Sligar, Y. Z. Ohkubo, and E. Tajkhorshid (2008) Blood clotting reactions on nanoscale phospholipid bilayers. Thrombosis Research, 122: S23-S26.
L. Thogersen, B. Schiott, T. Vosegaard, N. Chr. Nielsen, and E. Tajkhorshid (2008) Peptide Aggregation and Pore Formation in a Lipid Bilayer - a Combined Coarse Grained and All Atom Molecular Dynamics study. Biophysical Journal, 95: 4337-4347.
S. A. Shaikh and E. Tajkhorshid (2008) Potential cation and H+ binding sites in acid sensing ion channel-1. Biophysical Journal, 95: 5153-5164.
P.-C. Wen and E. Tajkhorshid (2008)
Dimer Opening of the Nucleotide Binding Domains of ABC Transporters after ATP hydrolysis.
Biophysical Journal, 95: 5100-5110.
(live molecular demo sessions of MalK dimers)
B. Isin, K. Schulten, E. Tajkhorshid, and I. Bahar. (2008) Mechanism of Signal Propagation upon Retinal Isomerization: Insights from Molecular Dynamics Simulations of Rhodopsin Restrained by Normal Modes. Biophysical Journal, 95: 789-803.
T. Vosegaard, K. Bertelsen, J. M. Pedersen, L. Thogersen, B. Schiott, E. Tajkhorshid, T. Skrydstrup, and N. Chr. Nielsen (2008) Resolution Enhancement in Solid-State NMR Spectra of Oriented Membrane Proteins by Anisotropic Differential Linebroadening. The Journal of the American Chemical Society, 130: 5028-5029.
2007
Y. Wang and E. Tajkhorshid (2007) Molecular mechanisms of conduction and selectivity in aquaporin water channels. Journal of Nutrition, 137: 1509S-1515S.
Y. Wang, J. Cohen, W. Boron, K. Schulten, and E. Tajkhorshid (2007) Exploring gas permeability of cellular membranes and membrane channels with molecular dynamics. Journal of Structural Biology, 157: 534-544.
M. Jensen, Y. Yin, E. Tajkhorshid, and K. Schulten (2007) Sugar transport across lactose permease probed by steered molecular dynamics. Biophysical Journal, 93: 92-102.
S. Tornroth-Horsefield, K. Hedfalk, M. Karlsson, U. Johanson, Y. Wang, E. Tajkhorshid, R. Horsefield, M. Nyblom, A. Backmark, P. Kjellbom, and R. Neutze (2007) Aquaporin gating. Journal of Biomolecular Structure and Dynamics, 24: 719-721.
2006
S. Tornroth-Horsefield, Y. Wang, K. Hedfalk, U. Johanson, M. Karlsson, E. Tajkhorshid*, R. Neutze*, and P. Kjellbom* (2006) Structural mechanism of plant aquaporin gating. Nature, 439: 688-694.
J. Yu, A. Yool, K. Schulten, and E. Tajkhorshid (2006) Mechanism of Gating and Ion Conductivity of a Possible Tetrameric Pore in Aquaporin-1. Structure, 14: 1411-1423.
F. Khalili-Araghi, E. Tajkhorshid, and K. Schulten (2006) Dynamics of K+ ion conduction through Kv1.2. Biophysical Journal, 91: L72-L74.
P. H. Koenig, N. Ghosh, M. Hoffmann, M. Elstner, E. Tajkhorshid, Th. Frauenheim and Q. Cui (2006) Towards theoretical analysis of long-range proton transfer kinetics in biomolecular pumps. Journal of Physical Chemistry A, 110: 548-563.
H.-C. Siebert, E. Tajkhorshid, J. F. G. Vliegenthart, C.-W. von der Lieth, S. Andre, and H.-J. Gabius (2006) Laser photo CIDNP technique as a versatile tool for structural analysis of inter- and intramolecular protein - carbohydrate interactions. In J. F. G. Vliegenthart, R. Woods (eds.), NMR spectroscopy and computer modeling of carbohydrates. ACS Symposia Series, 930: 81-113.
M. Hoffmann, M. Wanko, P. Strodel, P. H. Koenig, Th. Frauenheim, K. Schulten, W. Thiel, E. Tajkhorshid, and M. Elstner (2006) Color tuning in Rhodopsins: the mechanism for the spectral shift between bacteriorhodopsin and sensory rhodopsin II. Journal of the American Chemical Society, 128: 10808-10818.
Y. Yin, M. Jensen, E. Tajkhorshid, and K. Schulten (2006) Sugar binding and protein conformational changes in lactose permease. Biophysical Journal, 91: 3972-3985.